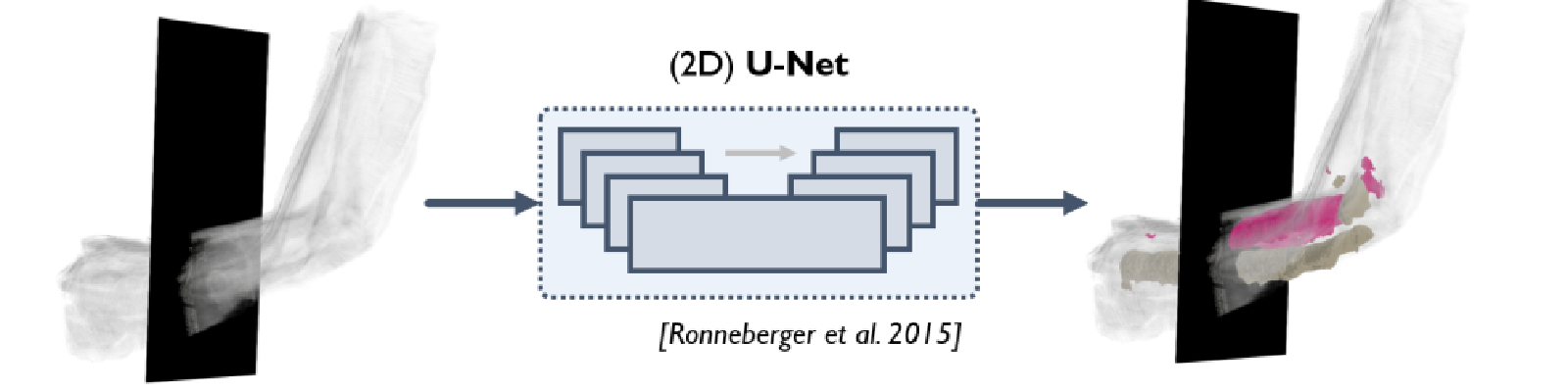
 Convolutional neural networks successfully annotate the biceps and humerus in motion-capture-localized ultrasound data.
Convolutional neural networks successfully annotate the biceps and humerus in motion-capture-localized ultrasound data.
OpenArm 2.0: Automated Segmentation of 3D Tissue Structures for Multi-Subject Study of Muscle Deformation Dynamics
 Convolutional neural networks successfully annotate the biceps and humerus in motion-capture-localized ultrasound data.
Convolutional neural networks successfully annotate the biceps and humerus in motion-capture-localized ultrasound data.
OpenArm 2.0: Automated Segmentation of 3D Tissue Structures for Multi-Subject Study of Muscle Deformation Dynamics
Abstract
We present a novel neural-network-based pipeline for segmentation of 3D muscle and bone structures from localized 2D ultrasound data of the human arm. Building from the U-Net neural network framework, we examine various data augmentation techniques and training data sets to both optimize the network’s performance on our data set and hypothesize strategies to better select training data, minimizing manual annotation time while maximizing performance. We then employ this pipeline to generate the OpenArm 2.0 data set, the first factorial set of multi-subject, multi-angle, multi-force scans of the arm with full volumetric annotation of the biceps and humerus. This data set has been made available on SimTK (https://simtk.org/projects/openarm) to enable future exploration of muscle force modeling, improved musculoskeletal graphics, and assistive device control.