The OpenArm Project:
Exploring Deformation as a Measure of Muscle Force
Laura A. Hallock and Ruzena Bajcsy
Human-Assistive Robotic Technologies (HART) Lab, University of California, Berkeley
Overview
While many musculoskeletal simulation frameworks rely on estimates of muscle force, there exists no noninvasive, in vivo method of measuring the force exerted by individual muscles. Such a measure would admit not only improved understanding of motor control and dexterity, but enhanced assistive device control and diagnosis and treatment of pathology.
We aim to address this force inference challenge by measuring muscle deformation, a signal that is intrinsically coupled to force production: as muscles shorten and widen during the cross-bridge cycle, tendons lengthen and force increases. This signal can be highly localized — allowing for insights related to individual muscles — but is also complex, requiring examination in many spatial and temporal dimensions and alongside activation measures like surface electromyography (sEMG) to gain phenomenological insight on the force–deformation relationship.
The OpenArm project comprises a number of investigations that address these challenges, each of which is detailed below. We invite you to peruse each of these subprojects and to make use of their associated open-source code and data sets for your own applications in biomechanics research, computer graphics, assistive device development, and beyond.
Time Series Force–Deformation–Activation Correlation
While the core mechanism relating muscle force to deformation is straightforward, geometric complexities and morphological differences across individuals mean that no generalizable force–deformation model yet exists. We show that simple measures of deformation of the brachioradialis muscle (including cross-sectional area, thickness, and aspect ratio), as measured via 2D ultrasound, correlate well with elbow output force and sEMG-measured activation under varied isometric contraction. These correlations vary both quantitatively and qualitatively by subject and by kinematic configuration of the elbow.
We are currently working to expand our analyses to more sophisticated deformation signals (e.g., statistical shape modeling, as enabled by the contour tracking work below) and to construct a testing platform enabling real-time, synchronized, simultaneous collection of ultrasound, sEMG, and force data for use in enhanced modeling and assistive device control.
All data and analysis code have been made available for general research use at the links below.
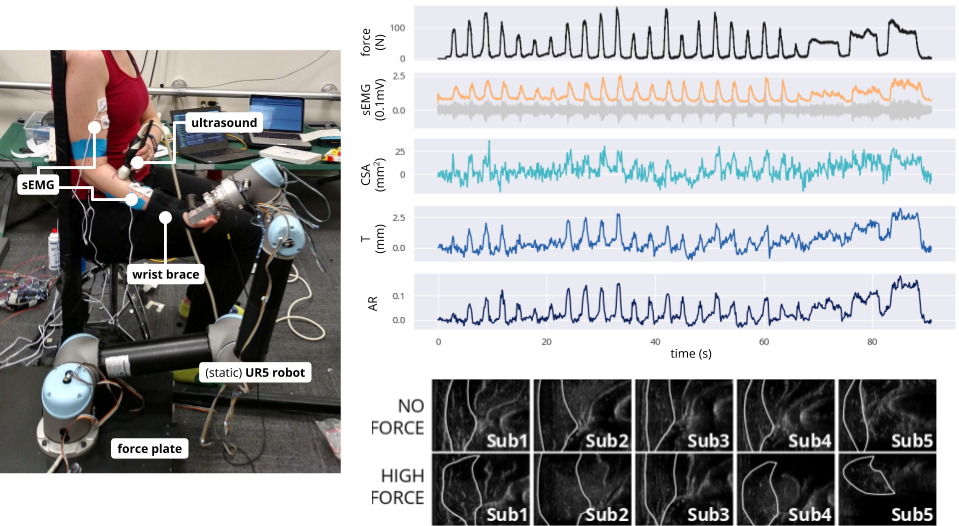
Left: Experimental platform allows for simultaneous measurement of muscle deformation, activation, and output force via ultrasound, surface electromyography (sEMG), and force plate, respectively, during varied isometric contraction of the elbow. Right, top: Time series data during single flexion trial show substantial correlation between force, sEMG, and muscle deformation (as cross-sectional area, thickness, and aspect ratio change of the brachioradialis). Right, bottom: Deformation of the brachioradialis cross section differs qualitatively across subjects, but reliably changes with force output.
Publication OpenArm Multisensor Data Set Import & Analysis CodeMuscle Contour Tracking
For muscle deformation to become a viable control signal (or even to be studied at scale without the need for extensive manual annotation), signals of interest must be tracked in an automated manner. We show that optical-flow-based methods allow for tracking of the brachioradalis contour, especially when combined with bilateral filtering and supporter-based methods, enabling real-time extraction of both the simple deformation signals explored in the above correlation studies (cross-sectional area, etc.) and more sophisticated ones (statistical shape models, dense motion analyses, etc.).
Optical flow tracking code, as well as data sets on which it can be applied, have been made available for general research use at the links below.
![]()
Brachioradialis contour, as tracked via variants of Lucas–Kanade optical flow. Left: Bilaterally-filtered Lucas–Kanade (BFLK) algorithm tracks much of the image contour reliably through fine and coarse filtering of each image frame (to track red and yellow points, respectively), but is susceptible to drift in areas of narrow fascia (e.g., upper right quadrant). Right: Supporter-based Lucas–Kanade (SBLK) algorithm tracks points along narrow fascia (green, closed) via "supporter" points (green, open) throughout the image, provided their motion is reliably correlated.
Publication OpenArm Multisensor Data Set Tracking CodeMuscle Force Modeling
While the simple measures of muscle deformation explored above correlate well with overall joint output force, deformation can be parameterized in a multitude of ways, and it is unclear a priori for which measures this correlation is most reliable. Choosing a single force–deformation modeling framework represents a fundamental trade-off: relying exclusively on empirical data (e.g., through a neural network model) leaves us with little scientific insight or generalizability, while choosing a physiological model assumes away significant geometric, mechanical, and neurological complexity that may prove critical to modeling success.
To balance these challenges, we propose a principled suite of models that make varying trade-offs between collected data and simplifying assumptions from literature in a quantifiable manner, ranging from purely data-driven "black box" to highly structured "white box" models. We are currently working to train and evaluate these models on the various 2D and 3D OpenArm deformation data sets above.
Preliminary, unpublished modeling work can be found at the link below.
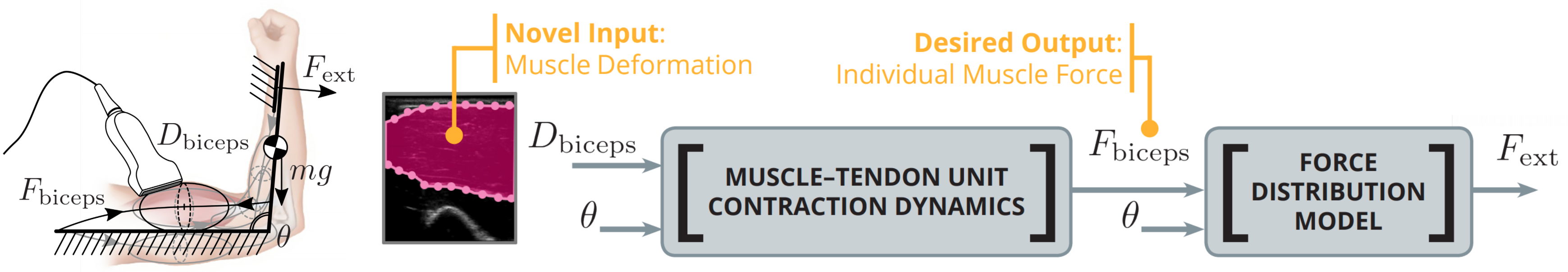
Proposed modeling framework by which muscle force can be inferred from various candidate deformation signals. See link below for details.
Preliminary Framework & Results3D Volumetric Data Generation & Analysis
Because the 2D-ultrasound-based deformation signals explored in previous correlation studies are complicated by geometric irregularities, changes in kinematic configuration, and contact dynamics with the surrounding tissues, a principled understanding of the force–deformation relationship requires examining the structures in 3D. We employed ultrasound and motion capture to generate full 3D volumetric data of the biceps brachii and surrounding tissue structures under multiple (static) elbow angles and force conditions. Preliminary analyses of these data sets — which have been made available for general research use as OpenArm 1.0 and 2.0 at the link below — indicate that force- and kinematic-associated deformation are qualitatively distinct and that the appearance of this deformation in cross section varies substantially based on spatial location along the arm.
Summary of data collection and preliminary analysis.
Publication OpenArm 1.0 & 2.0 Data SetsCNN-based Segmentation
Manual annotation of volumetric tissue structures (including muscles and bones) is prohibitively time-intensive, especially when these structures must be annotated at multiple time steps, as in the studies above. We developed a novel neural-network-based pipeline to segment 3D muscle and bone structures from the motion-capture-localized 2D ultrasound data generated in the studies above, enabling expansion of our annotated data sets to a larger cohort of subjects.
All neural network models, code, and generated annotated data sets (included in OpenArm 2.0 release) have been made available for general research use at the links below.
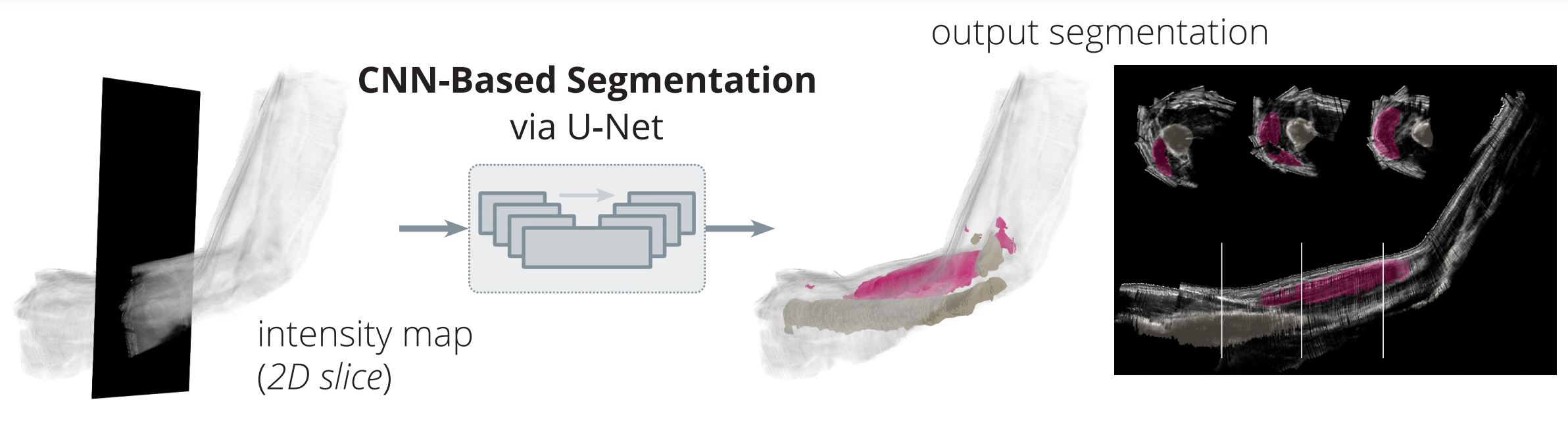
Convolutional neural networks successfully annotate the biceps and humerus in each slice of motion-capture-localized ultrasound data.
Publication Data Sets & Network Models Annotation CodePublication List
Laura A. Hallock, Akash Velu, Amanda Schwartz, and Ruzena Bajcsy. "Muscle Deformation Correlates with Output Force during Isometric Contraction." In BioRob, 2020.
Yonatan Nozik*, Laura A. Hallock*, Daniel Ho, Sai Mandava, Chris Mitchell, Thomas Hui Li, and Ruzena Bajcsy. "OpenArm 2.0: Automated Segmentation of 3D Tissue Structures for Multi-Subject Study of Muscle Deformation Dynamics." In EMBC, 2019. *Equal contribution.
Laura A. Hallock, Akira Kato, and Ruzena Bajcsy. "Empirical Quantification and Modeling of Muscle Deformation: Toward Ultrasound-Driven Assistive Device Control." In ICRA, 2018.
All PublicationsAcknowledgments & Sponsors
The authors acknowledge the aid and advice of Akash Velu, Amanda Schwartz, Akira Kato, Gregorij Kurillo, Yonatan Nozik, Daisuke Kaneishi, Robert Matthew, Sarah Seko, Daniel Ho, Sai Mandava, Chris Mitchell, Thomas Hui Li, Jeffrey Zhang, Ian McDonald, Sathvik Nair, Logan Howard, Yannan Tuo, and Vijay Govindarajan.
This work was supported by the NSF National Robotics Initiative (award no. 81774), Siemens Healthcare (85993), the NVIDIA Corporation GPU Grant Program, eZono AG, and the NSF Graduate Research Fellowship Program.
Thanks to Heidelberg University and MPIA for the multimedia poster template.